tDRnamer web server
The tDRnamer web server allows researchers to name tDRs by providing sequences or retrieve sequences by providing tDR names associated with model organisms. tDR sequence and tDR name examples which only take several seconds to run are available on the web server for demonstrating the functionality. The expected results generated by the tDRnamer web server are provided below. Please contact us to request for additional genome.
How to search by tDR sequences
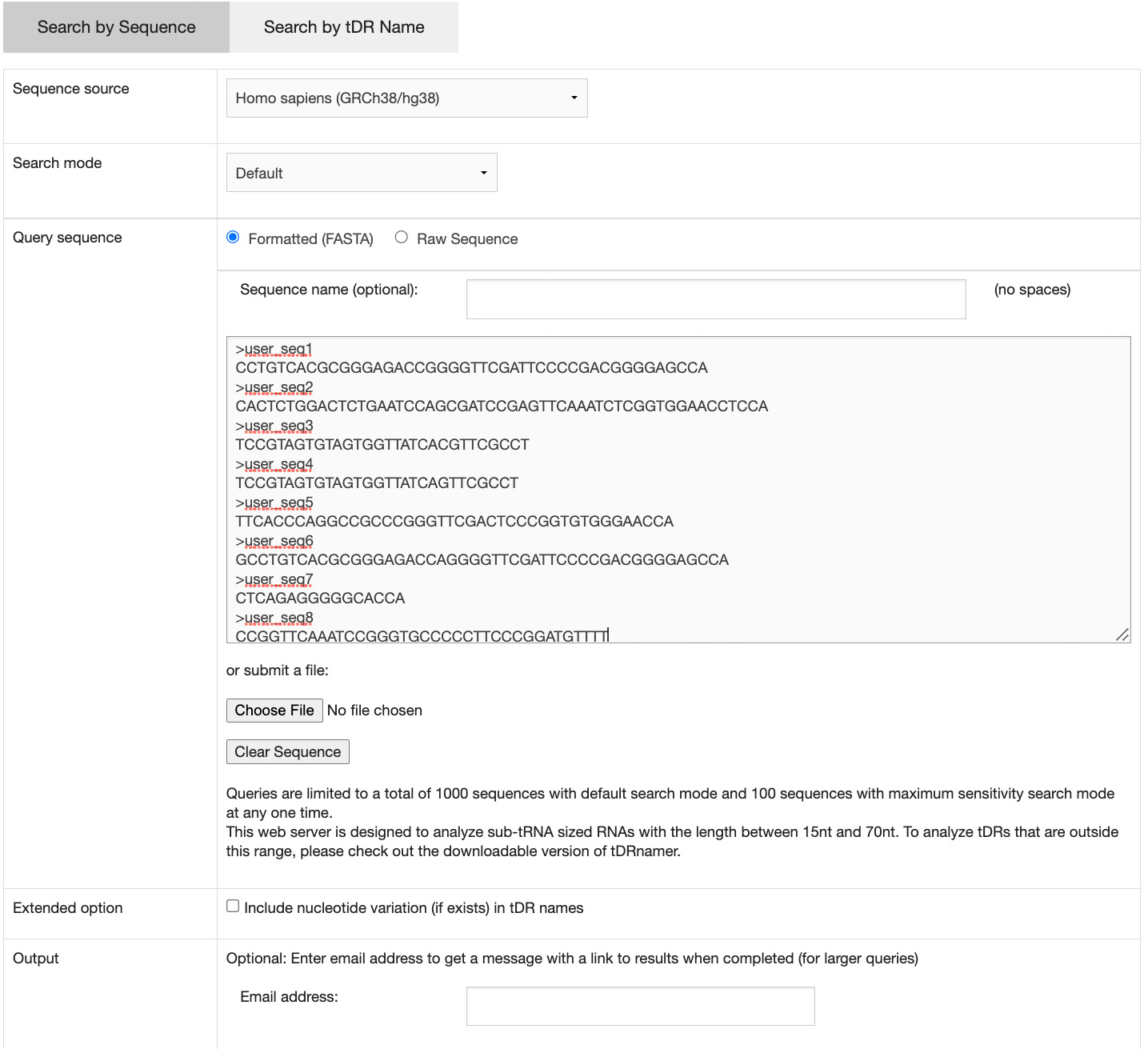
Clicking on the "Search by Sequence" tab will bring up the input form. There are two required items for initiating the search:
- sequence source - the genome assembly where the tDR sequences are originated
- query sequence - the tDR sequences can be entered in the text field or submitted by uploading a FASTA file
A maximum of 1000 sequences can be queried at one time with default search mode. Optionally, researchers can change the search mode to maximum sensitivity. However, due to the extended search time, the maximum number of queried sequences with maximum sensitivity search mode is 100. If more than 50 sequences are submitted, an email address is required to be provided for receiving a link to the results when the process is completed.
Both the forward and reverse strands of query sequences are searched.
By default, the names provided for the tDRs do not include the annotation of nucleotide variations found in the sequences. To include them as part of the tDR names, researchers can check the corresponding box on the search form.
How to search by tDR names
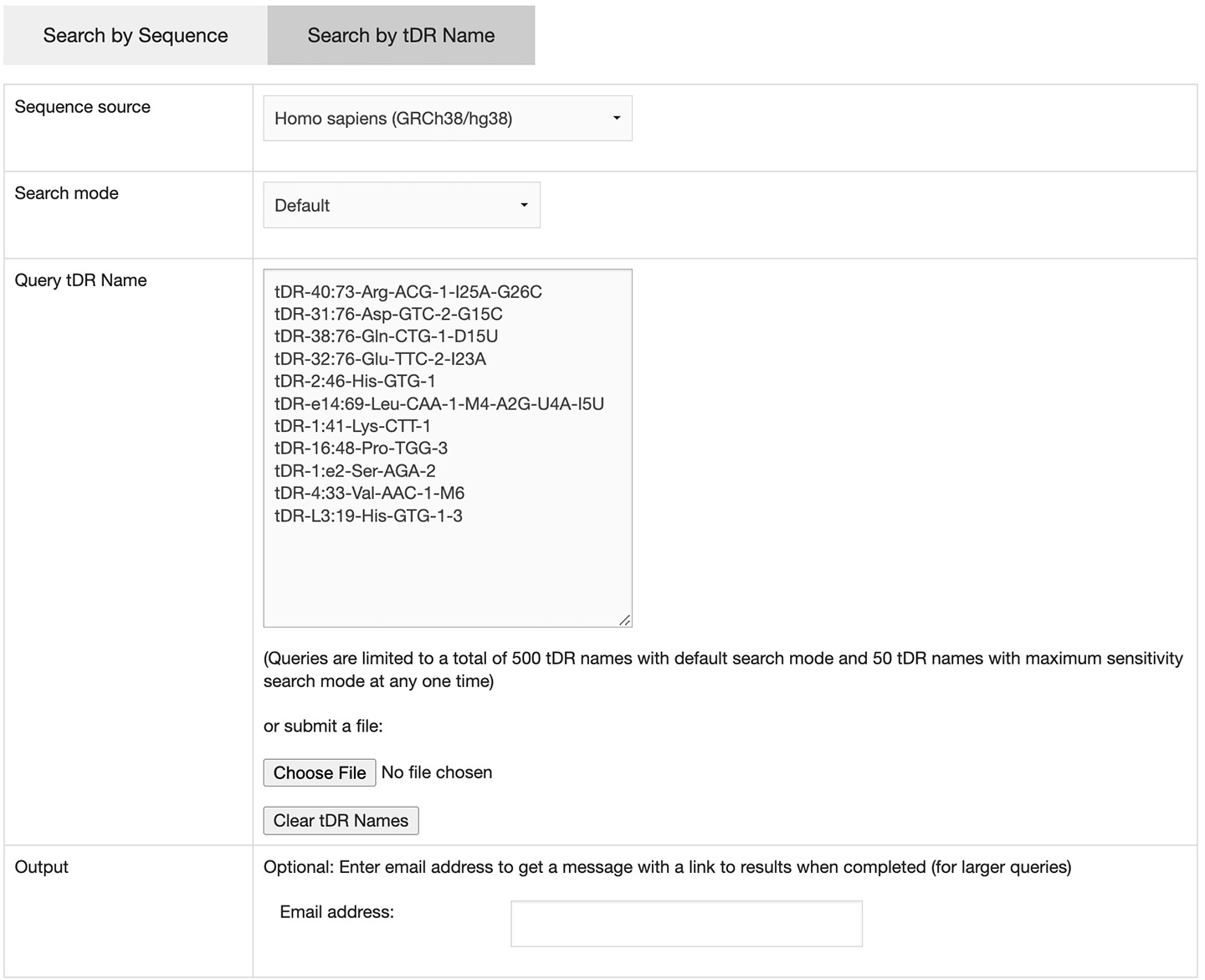
Clicking on the "Search by tDR Name" tab will bring up the input form. There are two required items for initiating the search:
- sequence source - the genome assembly where the tDRs are originated
- query tDR name - the tDR names formatted according to the our standardized naming system can be entered in the text field or submitted by uploading a text file
A maximum of 500 tDR names can be queried at one time. Optionally, researchers can change the search mode to maximum sensitivity. However, due to the extended search time, the maximum number of queried names with maximum sensitivity search mode is 50. If more than 50 names are submitted, an email address is required to be provided for receiving a link to the results when the process is completed.
Exploring tDR names and annotations
tDR naming and annotations are shown in three sections:
Summary
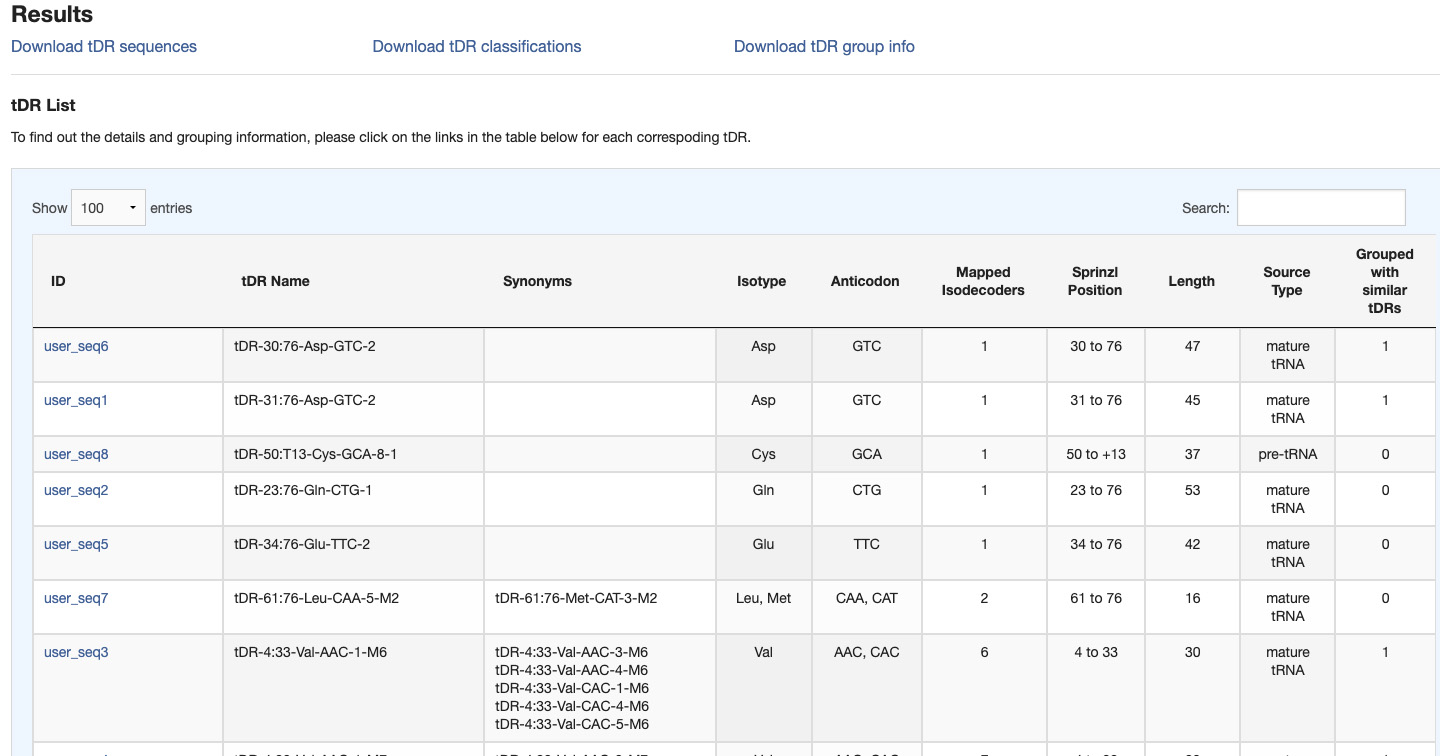
Results of the tDR annotations can be downloaded in text format by using the provided links.
- tDR sequences - FASTA file with tDR names and sequences
- tDR classifications - tab-delimited file with tDR annotations including tDR names and sequences, source tRNAs, positions of tDRs relative to source tRNAs, original tRNA isotype and anticodon, sequence variation counts, and group ID
- tDR group info - text file containing queried tDRs that are grouped together by sequence alignments
Information of queried tDRs are summarized in a table. They include:
- ID - sequence ID provided as inputs when searching by sequences
- tDR Name - standardized tDR name assigned to a corresponding queried sequence
- Sequence - tDR sequence when searching by tDR names
- Synonyms - other tDR names that correspond to the same tDR sequence
- Isotype - tRNA isotype of source tRNA(s)
- Anticodon - anticodon of source tRNA(s)
- Mapped Isodecoders - number of unique tRNA transcripts from which the tDR may derive
- Sprinzl Position - location of tDR relative to source tRNA(s)
- Length - length of tDR sequence
- Source Type - pre-tRNA or mature tRNA as tDR source
- Grouped with similar tDRs - number of other tDRs that can be grouped together by sequence alignments
tDR details
Clicking the link at the sequence ID in the result summary table launches the detail information of the corresponding tDR.
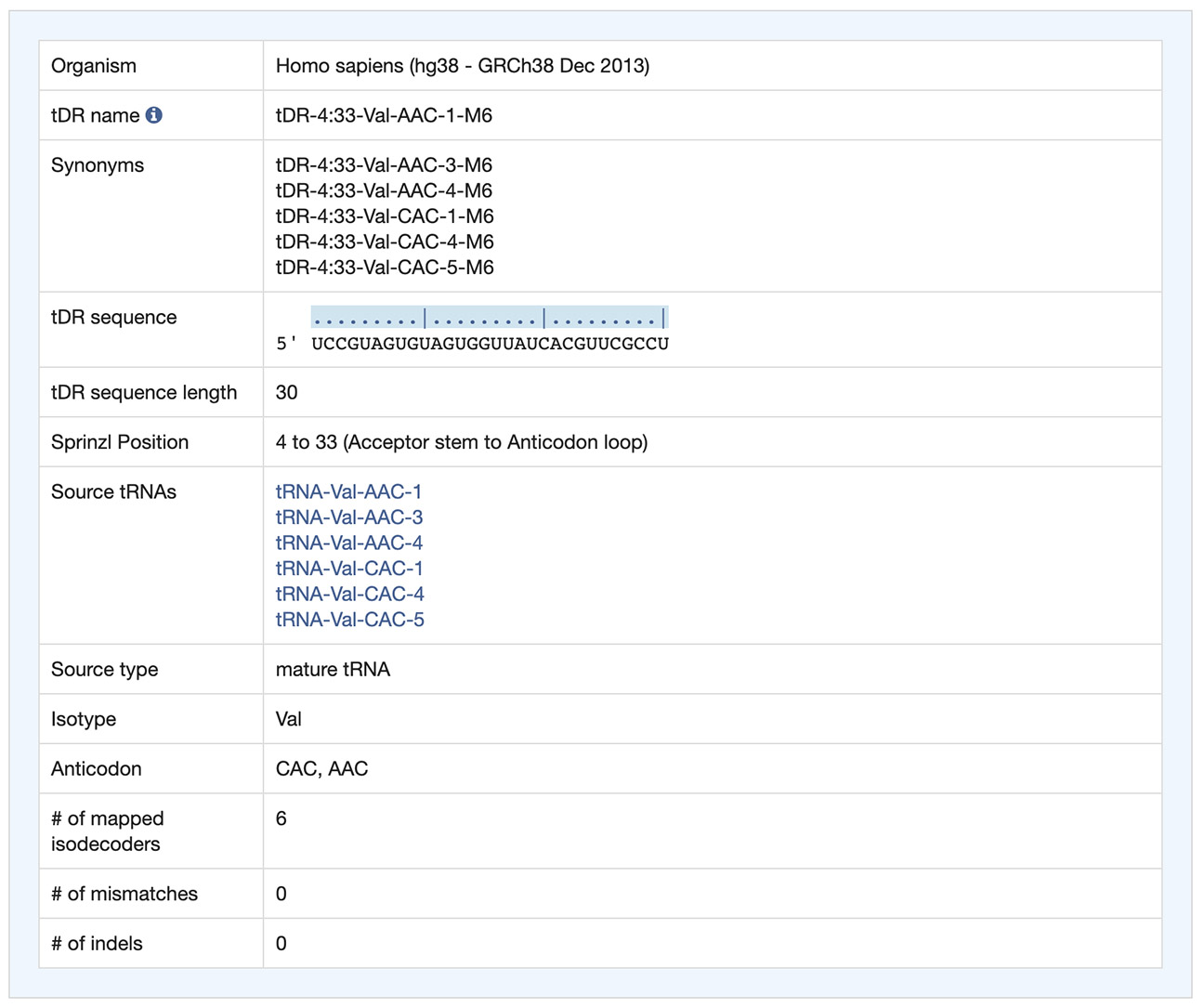
The overview information includes the tDR name and sequence, location of the tDR relative to source tRNAs, the source tRNA names with links to GtRNAdb, and any sequence variations that may exist based on alignments with source tRNAs.

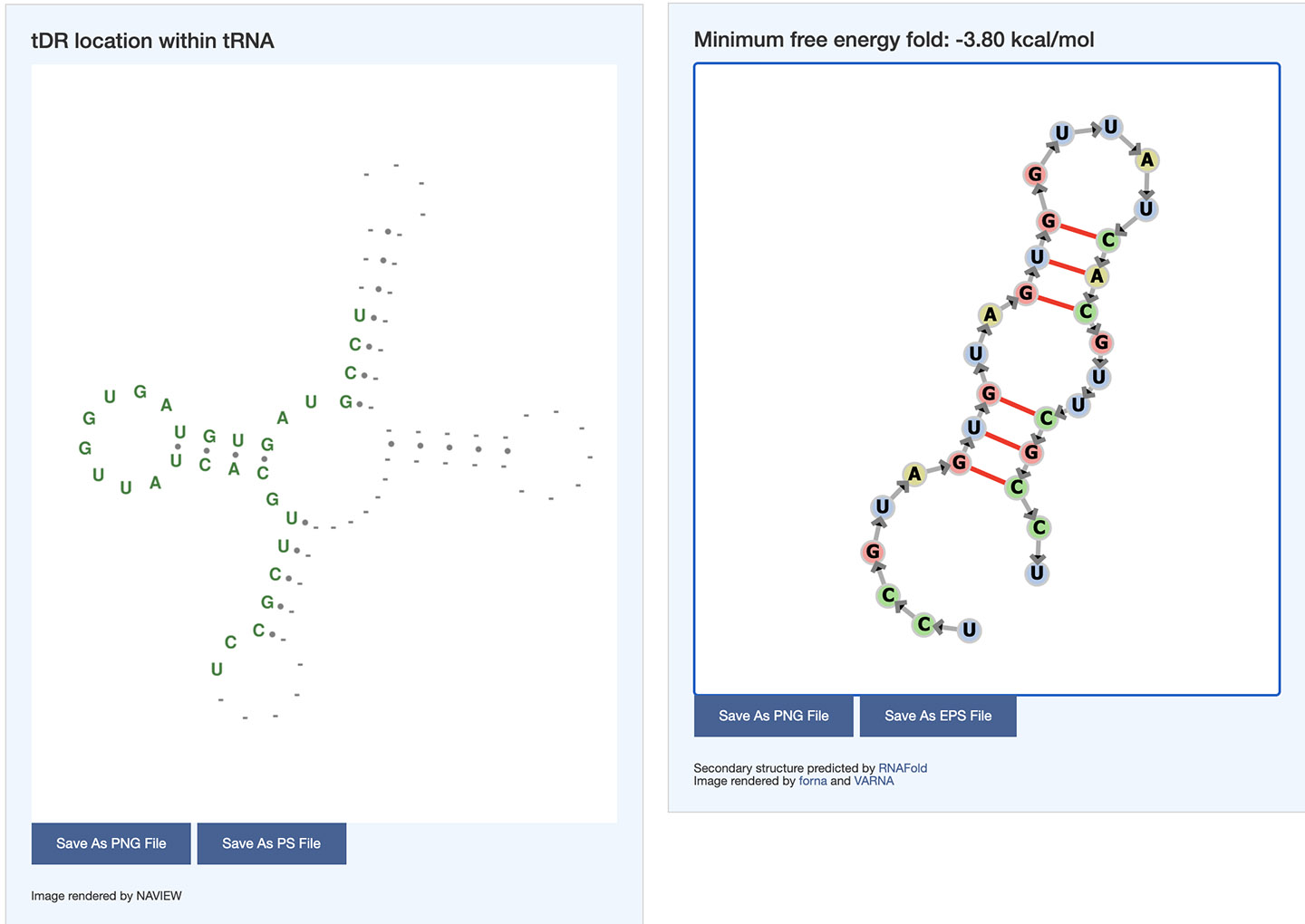
The alignments of the tDR sequence with the source tRNAs show the location of the tDR relative to the tRNA secondary structure. Nucleotide differences with source tRNAs are highlighted in red. Researchers can further visualize the tDR location in the superimposed tRNA secondary structure image. The secondary structure of the tDR was also predicted using minimum free energy method. The images can be saved in PNG and postscript file format.
tDR group
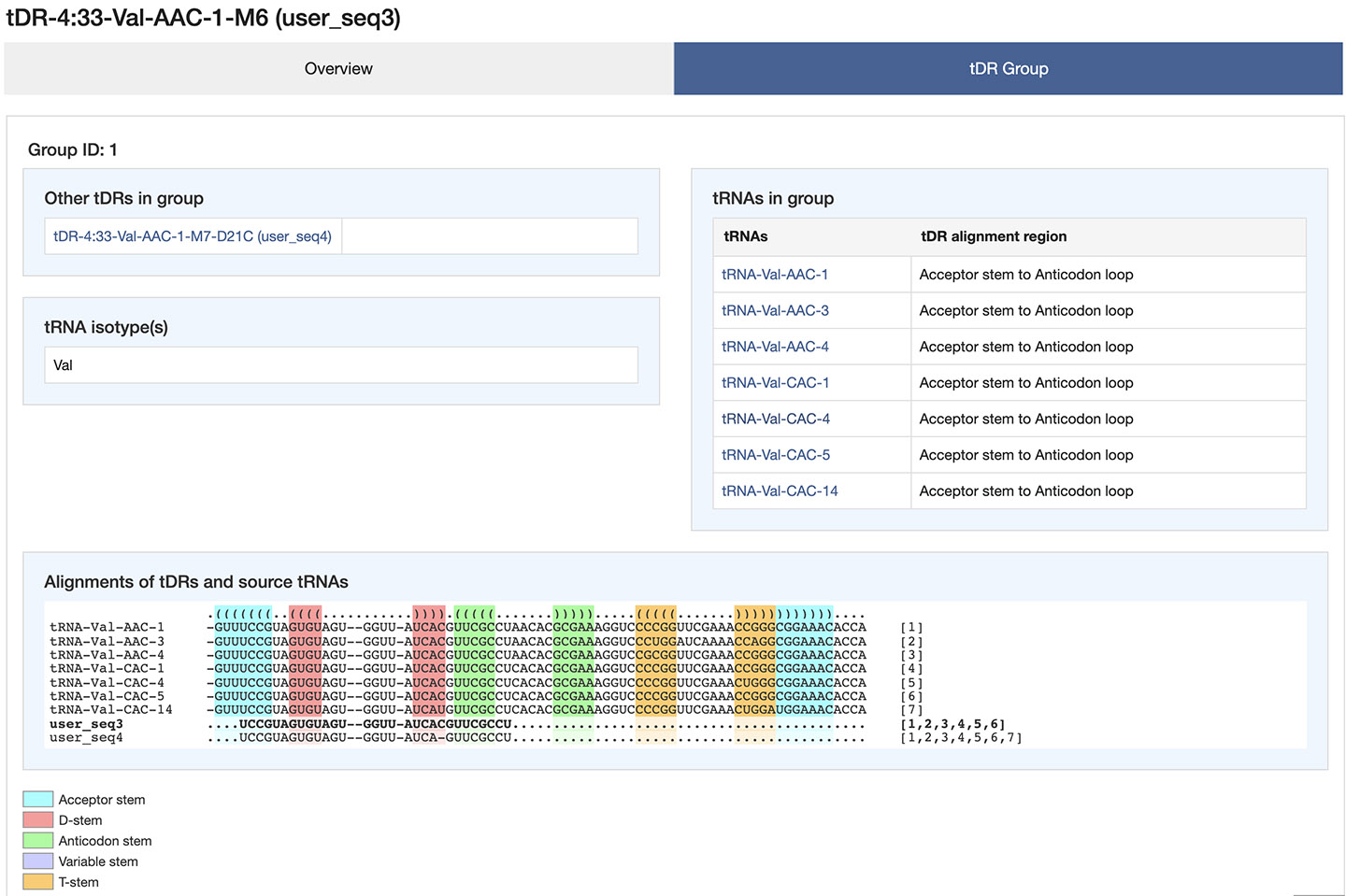
The tDR group includes all other queried tDRs that belong to the same group based on sequence alignments. Those tDRs can be viewed by clicking on the links in the list. Alignments of source tRNAs and tDRs in the group are provided with tRNA secondary structure annotations. The selected tDR is highlighted in the alignments. The numbers following the sequence of each tDR represent the corresponding source tRNAs in the alignments. Researchers can also obtain information of the source tRNAs by clicking on the corresponding links that launch the page in GtRNAdb.